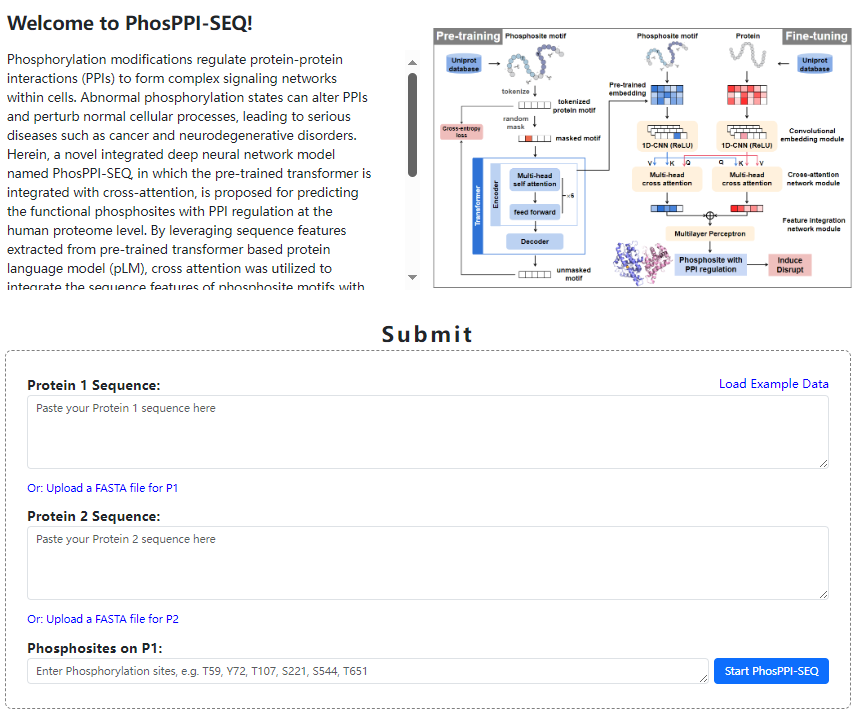
Introduction
The functional phosphosites for PPI regulation were collected from PhosphoSitePlus (PSP) and PTMint databases. Herein, we introduce a web server named PhosPPI-SEQ (Sequence-based Predictor for Phosphosites Regulating PPIs), which features a user-friendly interface. It can be used to assign PPI-related functions to phosphosites in proteomics. PhosPPI-SEQ can predict the functional scores for PPI regulation and the regulatory effects (induce or disrupt) of the functional phosphosites respectively. In this novel integrated deep neural network model, by leveraging the sequence features extracted from the pre-trained Transformer-based protein language model (pLM), the cross-attention mechanism was used to integrate the sequence features of phosphosite motifs and interacting proteins. This enables PhosPPI-SEQ to capture the complex interactions between functional phosphosites and interacting proteins and make judgments on the regulatory effects of sites with PPI regulatory capabilities.
Software Requirements
The PhosPPI-SEQ requires a modern web browser with JavaScript and cookies enabled. The following browsers have passed the PhosPPI-SEQ test:
- Mozilla Firefox, version 4 or above
- Chrome, version 5 or above
- The latest version of Firefox and Chrome is recommended for visualization.
Tutorials
This tutorial shows the user how to use the PhosPPI-SEQ to predict phosphosite function.
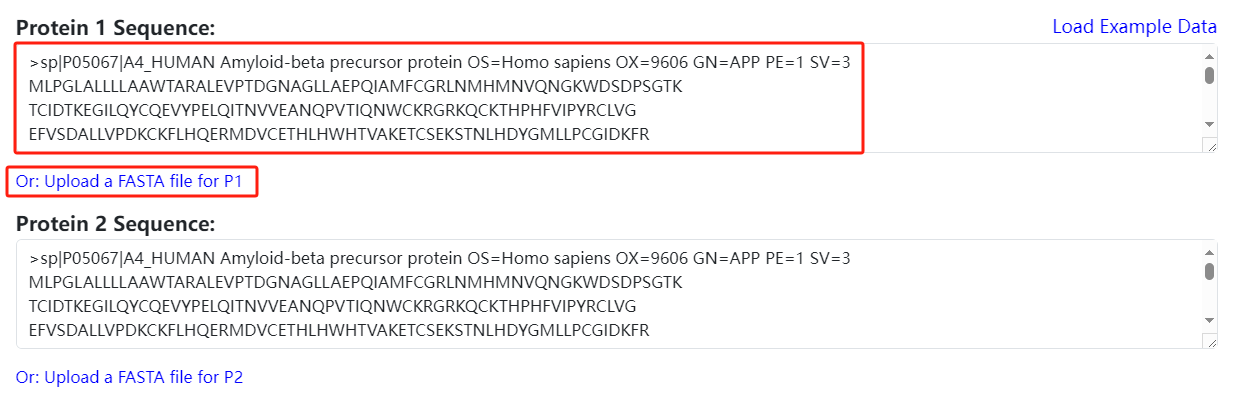
- Go to the prediction page: Click on Tools → PhosPPI-SEQ to enter the function prediction page.
- Input protein sequences: Two input methods are provided: text box input or loading a sequence fasta file (please use the standard format).
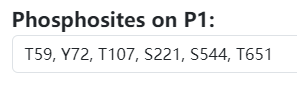
- Input phosphosites: Based on the protein information, users need to manually enter one or more phosphosites they wish to predict. The input format of phosphosites should include the residue type and sequence number. For example, in the given case, it could be Y169, Y580, T222, S18, T307, S621. Note that you can input phosphosites with residue types of S (serine), T (threonine), or Y (tyrosine) all at once, and the backend will automatically determine which corresponding models to be employed.
- Start prediction: Click the “Start PhosPPI-SEQ” button to predict the PPI-related functions of the phosphosites and their regulatory effects on PPIs.

Results
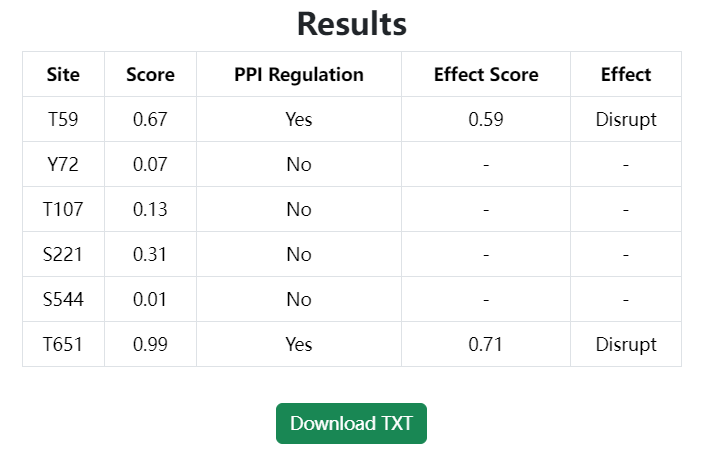
The forecast results are presented in tabular form.
Interpretation of Results:
- Site is the target phosphosite input into the model.
- Score and PPI Regulation indicate whether the phosphosite is likely to regulate PPI. Specifically, a Score > 0.5 indicates that the phosphosite is predicted to regulate PPI, and the PPI regulation is labeled with “Yes”, and vice versa.
- Effect Score and Effect reflect the specific regulatory effect of the phosphosite on PPI, only for the phosphosites with PPI regulation as “Yes”. Specifically, an Effect Score > 0.5 suggests that the phosphosite probably have a disruptive effect on PPI, labeled as “Disrupt”, whereas an Effect Score <= 0.5 indicates that it may have an inductive effect on PPI, label as “Induce”.
Click the “Download TXT” button to download the predicted score and effect of the phosphosites.