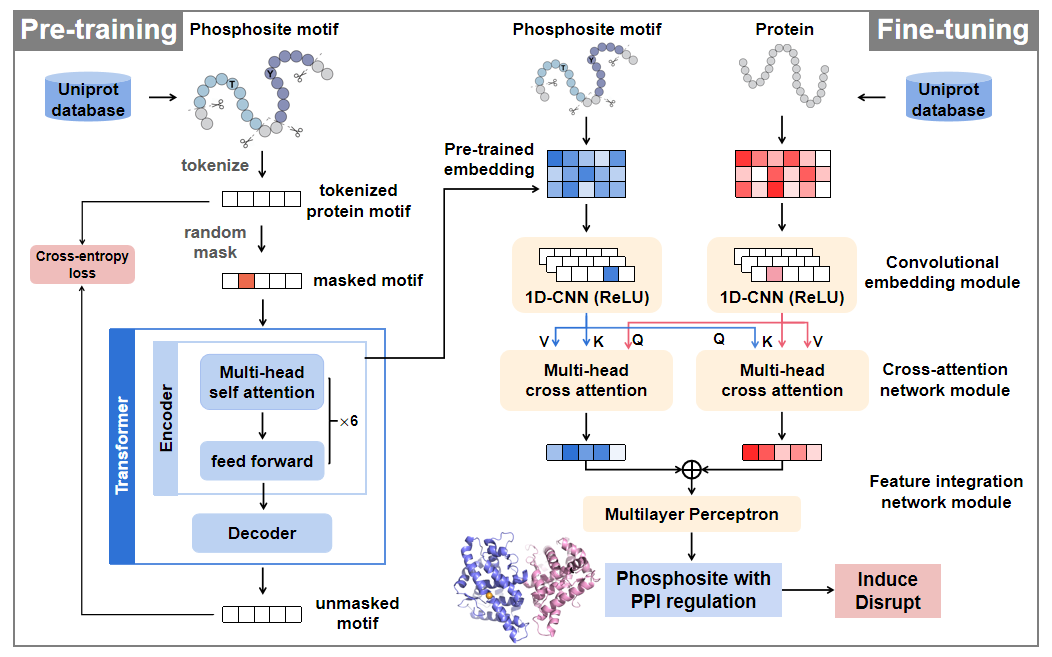
Welcome to PhosPPI-SEQ!
Phosphorylation modifications regulate protein-protein interactions (PPIs) to form complex
signaling networks within cells. Abnormal phosphorylation states can alter PPIs and perturb
normal cellular processes,
leading to serious diseases such as cancer and neurodegenerative disorders. Herein, a novel
integrated deep neural
network model named PhosPPI-SEQ, in which the pre-trained transformer is integrated with
cross-attention, is proposed for
predicting the functional phosphosites with PPI regulation at the human proteome level. By
leveraging sequence
features extracted from pre-trained transformer based protein language model (pLM), cross
attention was utilized to integrate
the sequence features of phosphosite motifs with interacting proteins, allowing PhosPPI-SEQ
to
capture the complex
interactions between the functional phosphosites and interacting proteins.
PhosPPI-SEQ requires only the raw protein sequences of the concerned PPI, and concerned
phosphosites as input, avoiding
the problem of losing information for a large amount of phosphosites due to the limited
number
of protein crystal
structures. It is computationally efficient and has a broader scope of application for
biologists.